Targeting Mutational Landscape of TP53 in patients diagnosed with Oral Cancer living in Senegal
Abstract
Introduction
Genomic mutations in TP53 gene in association with etiological risk factors have been associated with oral carcinogenesis. Herein, we screened for genomic variants of TP53 predisposing to oral cancers in Senegalese patients.
Methodology
88 patients with confirmed diagnostic were recruited after informed consent. Blood samples were collected from each patient to perform DNA extraction, PCR amplification of all coding exons of TP53 followed by Sanger Sequencing of PCR products. Nucleotide sequences were analysed with Genalys software. 94 blood donors with no cancer diagnosis were also recruited as controls for association study between the most common variants identified in patients and predisposition to oral cancers.
Results
Sequence analysis showed that 52.27% of patients carry at least one mutation in TP53. Eleven genomic variants were identified, 7 variants already reported in databases and 4 new variants. The most recurrent variants in this study already reported as cancer-related variants were Pro72Arg (rs1042522; Arginine frequency estimated at 31.26%) and a 16 bp insertion in intron 3 (rs59758982; allelic frequency estimated at 26.25%). Haplotype analysis between these variants showed a strong linkage disequilibrium (D’ = 0.999, r2 = 0.153 and p-value < 0.05). However, association study did not find any significant association with susceptibility to oral cancer (p-value > 0.05).
Conclusion
Our study highlighted that despite the absence of association between the two most common cancer-related variants in Senegalese patients diagnosed with oral cancer, their strong LD suggested that they could be transmitted together in a common haplotype which may be implicated in oral carcinogenesis.
Author Contributions
Academic Editor: Qingwen Xu, United States.
Checked for plagiarism: Yes
Review by: Single-blind
Copyright © 2022 SARR Pierre Diaga, et al.
 This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Competing interests
The authors have declared that no competing interests exist.
Citation:
Introduction
With an estimated incidence of nearly 300.000 new cases per year and a mortality rate around 50%, oral cancers are major challenge in oncology and represent the sixth leading cancer worldwide, according to GLOBOCAN report 2018 1.
The main risk factors are exposure to genotoxic agents such as tobacco, alcohol and betel nut chewing 2, 3, 4.These agents induced genetic alteration in genes controlling the cell cycle such as TP535, 6, 7. Also, poor oral hygiene, bad set of teeth, inadequate diet and poor nutrition, or immunosuppression, may increase oral cancer risk by promoting chronic infections of the oral mucosa with high risk Human Papilloma Virus (HPV), by inducing loss of function of TP53 encoded protein 8, 9, 10.
Mutations of TP53 gene have been reported as the main driver event of carcinogenesis 11. Missense variants have been reported in the coding region of the DNA Binding Domain of p53 protein which specifically binds to the promoters of targeted genes.
Eight hotspot amino-acid substitutions in the DNA Binding Domain of p53, characterizing almost 27% of mutant proteins, were identified in human cancers while they are not directly cancer associated (Arg175His, Gly245Ser, Arg248Gln, Arg248Trp, Arg249Ser, Arg273His, Arg273Ser, and Arg282Trp) 12. In mice model, the introduction of the Arg172His mutation corresponding to human Arg175His, has been associated with the functional inactivation of p63 and p73 proteins 13, 12.
Other variants are located in a 1kb region spanning intron 3 to intron 4 containing the 6 cancer-related variants reported in IARC TP53 database 14: a 16 bp insertion in intron 3 (NG_017013.2:g.16185_16200Ins, rs59758982), two variants located in exon 4 coding for the Proline-rich domain of p53 (Pro47Ser : NG_017013.2:g.16321C>T, rs1800371) and Pro72Arg : NG_017013.2:g.16397C>G, (rs1042522)), and 2 variants located in intron 4 (NG_017013.2:g.17032T>C (rs1794287) and NG_017013.2:g.17198G>A (rs35850753) (Figure 1).
Figure 1.Cancer-related variants of TP53 encompassing exons 4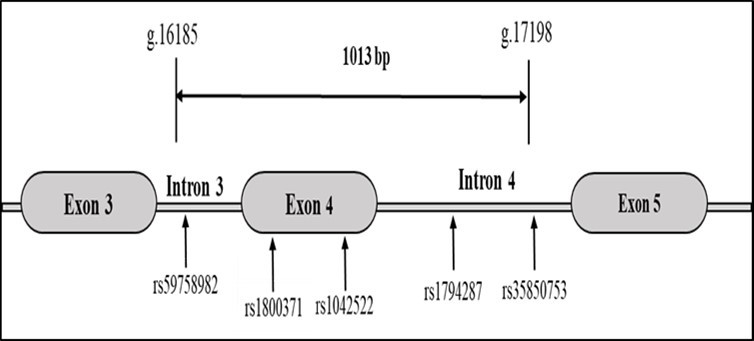
Effect predictions of these cancer related variants in Varsome database have shown benign effect (rs1800371, rs78378222 and rs35850753) or uncertain significance (rs59758982, rs1042522 and rs1794287), although functional/association studies are in favour of cancer predisposition (Table 1).
Table 1. VarSome effect predictions and functional study findings on cancer-related variants reported in IARC TP53 database| Genetic variations | Location | rs number | Effect Prediction | Functional/Association study findings |
| NG_017013.2:g.16185_16200Ins | Intron 3 | rs59758982 | VUS | Association with increased risk of colorectal cancer and reduced level of TP53 mRNA in lymphoblastoïd cell-lines [15] |
| NG_017013.2:g.16321C>T(p.Pro47Ser) | Exon 4 | rs1800371 | Benign | Association with breast cancer risk [17] |
| NG_017013.2:g.16397C>G(p.Pro72Arg) | Exon 4 | rs1042522 | VUS | Association with altered electrophoretic mobility and, a conformational and functional modifications of the p53mutant protein [18] |
| NG_017013.2:g.17032T>C | intron 4 | rs1794287 | VUS | Affect the activity of TP53 internal promoter [20] |
| NG_017013.2:g.17198G>A | intron 4 | rs35850753 | Benign | Association with neuroblastoma [21] |
| NG_017013.2:g.24117A>C | 3’ UTR | rs78378222 | Benign | Association with neuroblastoma [21] |
The 16 bp insertion in intron 3 (rs59758982) has been associated with increased risk of colorectal cancer in a case-control study and correlated with a reduced level of TP53 mRNA in lymphoblastoid cell-lines 15. The Pro47Ser variant (rs1800371) has been reported in populations of African ancestry (allelic frequency estimated at 2-4% in Africa and 1.2% in African-Americans) 16 and have been associated with increased risk of breast cancer among pre-menopausal women 17. The Pro72Arg (rs1042522) is a non-conserved amino acid with reported altered electrophoretic mobility, conformation and function of themutant protein 18. This variant shows significant ethnic bias with Arginine allelic frequencies estimated at ~72% for Europeans compared to ~38% for African and African-Americans and ~51% for Asian populations (gnomAD database: https://gnomad.broadinstitute.org/variant/17-7579472-G-C?dataset=gnomad_r2_1). In a previous study, we showed in an ethnicity-stratified meta-analysis that this variant is associated with oral cancers risk in Asian population (OR = 1.31; CI95% =1.09 - 1.58; p=0.004) whereas any significant association was observed in Africans and Caucasians 19.
Two other cancer-related variants (rs1794287) and (rs35850753) are located in intron 4 and were shown to affect the activity of TP53 intron 4 internal promoter by changing its affinity with several transcription factors 20. The last variant (rs78378222) maps to the polyadenylation signal located in 3´ UTR of TP53. This variant is in linkage disequilibrium with rs35850753 in intron 4 and both have been associated with n euroblastoma (rs35850753: OR = 2.7, CI95% = (2.0 - 3.6); rs78378222: OR = 2.3, CI95% = (1.8 - 2.9) 21.
In Sub-Saharan Africa, very few studies have investigated genetic variability of TP53 and its association with cancer risk. Here we screened for TP53 genomic variants predisposing to oral cancers in patients living in Senegal.
Population and Methods
The study was carried out from January 2018 to September 2021 through a collaboration between the Department of Stomatology and Maxillofacial Surgery of Aristide Le Dantec Hospital in Dakar for patients recruitment, the Senegalese National Blood Transfusion Center for the recruitment of healthy blood donor controls, the department of Human Genetics of the Faculty of Medicine, Pharmacy and Odontology of Cheikh Anta Diop University Dakar for DNA extraction and PCR amplification, and the Functional Genomics Platform of the National Center for Scientific and Technical Research (CNRST) of Rabat (Morocco) for Sanger sequencing of PCR products. The research protocol was approved by the Ethics Committee of Cheikh Anta Diop University under reference 0320/2018/CER/UCAD.
Study Population
Patients with confirmed diagnosis of oral cancer undergoing treatment at the Department of Stomatology and Maxillofacial Surgery of Aristide Le Dantec Hospital were recruited after informed consent. A questionnaire was filled out by each patient providing information about age, gender, ethnicity, alcohol and tobacco status, tumor location and histology, and disease stage.
94 healthy blood donors without cancer were also recruited as controls. For each individual a blood sample was collected for DNA extraction.
DNA Isolation, PCR Amplification of TP53 Coding Exons
DNA was isolated from blood samples using Quick-DNA™ MiniPrep (Zymo Research) following manufacturer’s protocol. DNA extract were quantified with a spectrophotometer (SimpliNano™, Biochrom). PCR amplifications of the 10 coding exons (exons 2 to 11) of TP53 gene were processed with seven primer sets at specific annealing temperatures (Table 2). PCR conditions were as follow: initial denaturation at 95°C for 10 min; 40 cycles of denaturation at 94°C for 30s, primer annealing for 30s, and extension at 72°C for 30s. A final extension step followed at 72°C for 10 min. PCR products were visualized by electrophoresis in a 1.5% agarose gel.
Table 2. Primer sets and annealing temperature of TP53 coding exons| Fragments; length in pb | Sequence of Primer sets | Annealing temperature |
| Exon 2 | E2F : CAGCCATTCTTTTCCTGCTC | 620C |
| 355 | E2R : TCCCACAGGTCTCTGCTAGG | |
| Exons 3 & 4 | E3F : CCCCCTCTGAGTCAGGAAACA | 550C |
| 765 | E4R : ACAGGAGTCAGAGATCACACA | |
| Exons 5 & 6 | E5F : TGAGGTGTAGACGCCAACTCT | 550C |
| 668 | E6R : GGGAGGTCAAATAAGCAGCA | |
| Exon 7 | E7F : CTTGCCACAGGTCTCCCCAA | 600C |
| 237 | E7R : AGGGGTCAGCGGCAAGCAGA | |
| Exons 8 & 9 | E8F : TTGGGAGTAGATGGAGCCTG | 600C |
| 473 | E9R : AAACAGTCAAGAAGAAAACGGC | |
| Exon 10 | E10F : GCTGTATAGGTACTTGAAG | 550C |
| 343 | E10R : GCTTTCCAACCTAGGAAGGCAG | |
| Exon 11 | E11F : GATTTGAATTCCCGTTGTCC | 550C |
| 324 | E11R : CAAGGGTTCAAAGACCCAAA |
The 16 bp insertion in intron 3 was genotyped in control population by PCR amplification using forward (TGCTCTTGTCTTTCAGACTTCCT) and reverse primer (GAGCAGTCAGAGGACCAGGTC) at 62°C annealing temperature during 30s. PCR products were visualized in a 3% agarose gel after electrophoresis at 100 volts for an hour. Two fragments were observed (114bp for the insertion allele and 130bp for the wild type allele).
The Pro72Arg variant (c.215C>G) was genotyped in controls by PCR-RFLP as previously reported 22.
Sanger Sequencing of PCR Products and Sequence Analysis
PCR products were cleaned with PureLink™ Quick Gel Purification kit (Invitrogen) according to manufacturer’s protocol. Cleaned PCR products were sequenced with BigDye™ Terminator kit (Applied Biosystems) and loaded on a SeqStudio™ Genetic Analyzer System 4 capillary (Applied Biosystems).
Sequences analysis were performed with Genalys Software (version 3.3.42a) and mapped to the reference sequence of TP53 gene (NG_017013.2) for variants detection. Clinical significance of identified variants was checked on ClinVar, dbSNP, VarSome and gnomAD databases. For newly identified variants, we predicted their biological effects with SIFT and PolyPhen2 tools.
Statistical Analysis
Linkage Desequilibrium (LD) was tested with Haploview Software (version 4.1). The association between the most common variants and predisposition to oral cancers was checked in a case/control study. Association parameters (odds ratio (OR), 95% confidence interval (CI95%) and p-value) were estimated through an online dedicated website 23.
Results
Characteristics of Study Population
The study population included 88 patients diagnosed with oral cancers. Mean age at diagnosis was 51.90 ±17.98 years with ages ranging from 18 to 85 years. Sex ratio was in favor of females (0.76) and most of our patients (73%) had a negative alcohol-tobacco status. Tumor histology showed that 90.9% were squamous cell carcinoma. Tumors were preferentially located in the tongue (22.7%), followed by gum (20.5%), cheek mucosa (19.3%), facial massif (8%), lips (6.8%), oral floor (3.4%), palate (2.3%), mandible (2.3%) and maxillary (1.1%) (Table 3).
Table 3. Characteristics of recruited patients| Characteristics | Parameters | Values |
|---|---|---|
| Age | Mean age ± standard deviation | 51.90 ± 17.98 |
| Median age extreme values | 55 (18 – 85) | |
| Sex-ratio | 0.76 (38/50) | |
| Smoking and alcohol status | - | 73 % |
| + | 5.5 % | |
| Not specified | 21.5 % | |
| Tumor location | Tongue | 22.7 % |
| Gum | 20.5 % | |
| Cheek mucosa | 19.3 % | |
| Facial massif | 8 % | |
| Lips | 6.8 % | |
| Oral floor | 3.4 % | |
| Palate | 2.3 % | |
| Mandible | 2.3 % | |
| Maxillary | 1.1 % | |
| Multiple locations | 11.3 % | |
| Not specified | 2.3 % | |
| Tumor histology | Squamous cell carcinoma | 90.9 % |
| Adenocarcinoma | 1.1 % | |
| Burkitt lymphoma | 1.1 % | |
| Not specified | 6.9 % | |
TP53 Mutation Screening
The proportion of patients carrying at least one mutation in TP53 was 52.27%. Sequence analysis identified 11 genomic variations, 10 Single Nucleotide Variant (SNV) and one Copy Number Variation (CNV) located in intron 3. Among identified variants, seven have been reported in Clinvar, dbSNP or gnomAD databases: c.63C>T (p.Asp=, rs1800369); c.215C>G (p.Pro72Arg, rs1042522); c.692C>T (p.Thr231Ile, rs1555525564); c.745A>T (p.Arg249Trp, rs587782082); c.773A>T (p.Glu258Asp, rs2073239956); c.776A>T (p.Asp259Val, rs745425759) and the CNV (GGGCTGGGGACCTGGA, NG_017013.2:g.16185_16200Ins, rs59758982).
We have identified four new variants not yet reported in variant databases: c.62A>C (p.Asp21Ala); c.64C>G (p.Leu22Val); c.268T>A (p.Ser90Thr) and c.773A>T (p.Glu258Val). Each of them was detected in one patient in the study population. Effect prediction with SIFT and PolyPhen highlighted possibly damaging effect (c.62A>C (p.Asp21Ala); c.773A>T (p.Glu258Val)) and benign effect (c.64C>G (p.Leu22Val); c.268T>A (p.Ser90Thr) (Table 4).
Table 4. TP53 mutations identified in patients with oral cancer and their clinical significance| c(g)DNA description | rs number | Genomic location (GRCh38) | Exon/Intron location | Protein effect | Variant Clinical significance ( ClinVar , dbSNP & gnomAD ) vs. Effect Prediction (SIFT & PolyPhen ) | Alternative allele frequency |
| c.62A>C | --- | 17:7676533 | Exon 2 | p.Asp 21Ala | Possibly damaging ( PolyPhen ) | 0.57 % (01/88) |
| c.63C>T | rs1800369 | 17:7676532 | Exon 2 | p.Asp21= | Conflicting interpretations of pathogenicity (ClinVar & dbSNP) | 0.57 % (01/88) |
| c.64C>G | --- | 17:7676531 | Exon 2 | p.Leu 22Val | Benign ( PolyPhen ) | 0.57 % (01/88) |
| g.16185_16200Ins | rs59758982 | 17:7676351_7676366Ins | Intron 3 | --- | Benign ; Likely-benign ; Hereditary cancer-predisposing syndrome (ClinVar & dbSNP) | 26.25 % (18/40) |
| c.215C>G | rs1042522 | 17:7676154 | Exon 4 | p.Pro72Arg | Uncertain clinical significance ; pathogenic (ClinVar & dbSNP) ; Tolerated (SIFT) ; Benign (PolyPhen) | 31.26 % (23/48) |
| c.268T>A | --- | 17:7676101 | Exon 4 | p.Ser 90Thr | Benign ( PolyPhen ) | 1.02 % (01/49) |
| c.692C>T | rs1555525564 | 17:7674271 | Exon 7 | p.Thr231Ile | Uncertain clinical significance (ClinVar & dbSNP) ; Deleterious (SIFT) ; Probably damaging (PolyPhen) | 0.98 % (01/51) |
| c.745A>T | rs587782082 | 17:7674218 | Exon 7 | p.Arg249Trp | Uncertain clinical significance ; likely pathogenic ; Deleterious (SIFT) ; Possibly damaging (PolyPhen) | 2.94 % (03/51) |
| c.773A>T | --- | 17:7674190 | Exon 7 | p.Glu 258Val | Probably damaging ( PolyPhen ) | 0.98 % (01/51) |
| c.773A>T | rs2073239956 | 17:7674189 | Exon 7 | p.Glu258Asp | Clinical significance not reported in Clinvar & dbSNP ; Probably damaging (PolyPhen) | 1.96 % (02/51) |
| c.776A>T | rs745425759 | 17:7674187 | Exon 7 | p.Asp259Val | Uncertain significance (ClinVar & dbSNP) ; Deleterious (SIFT) ; Possibly damaging (PolyPhen) | 0.98 % (01/51) |
The most recurrent variants were two cancer-related variants reported in IARC TP53 database: p.Pro72Arg (rs1042522; Arginine allelic frequency estimated at 31.26%) and the CNV located in intron 3 (rs59758982; alternative allelic frequency estimated at 26.25%). Since these variants are located closely in the genomic region spanning intron 3 to exon 4, we then tested for linkage desequilibrium (LD) and found a strong LD (Table 5).
Table 5. Linkage desequilibrium calculation between the two most frequent variants| Markers | D | D’ | r 2 | p-value |
|---|---|---|---|---|
| rs59758982 vs. rs1042522 | -0.079 | 0.999 | 0.153 | 0.000551698 |
Case/controls association study between the two most frequent variants and oral cancer predisposition did not raise any significant association in the study population (Table 6).
Table 6. Case/control association study of rs59758982 and rs1042522 with predisposition to oral cancer| Variants | Allelic frequencies | Test of association | |
|---|---|---|---|
| Cases | Controls | ||
| 16 bp insertionrs59758982 | 26.25 % | 27.5 % | OR = 1.07CI 95% = (0.57 – 2)p = 0.841 |
| Arg allelers1042522 | 31.26 % | 32.8 % | OR = 0.93CI 95% = (0.51 – 1.68)p = 0,823 |
Discussion
Clinical characteristics of Senegalese patients diagnosed with oral cancers have been described by Touré et al in 2005 and highlighted early age at diagnosis (52.6 years), a sex ratio in favor of females (0.67), a negative alcohol-tobacco status (62.9% non-smokers and 83.8% non-alcoholics) and a poor oral hygiene 24. This profile is quite similar to what observed in our patients. These results highlighted that tobacco and alcohol are not the common risk factors and emphasized the hypothesis that exposure to HPV and genetic factors may be associated with oral carcinogenesis in Senegalese population.
The predominant tumor histology in our patients was squamous cell carcinoma. This type represents 95% of all oral cancers tumors worldwide and is characterized by altered proliferation of dysplastic squamous cells on the surface of the epithelial layer ) 12. In mice model, the introduction of the Arg172His mutation corresponding to human Arg175His, has been associated with the functional inactivation of p63 and p73 proteins 13, 12.
Among oral cancer etiological factors genetic alterations in specific genes controlling the cell cycle such as TP53 have been reported. In this study, 52.27% of patients carried at least one mutation in TP53 (46/88), similar to data from Poeta et al in the US (53.3%, 224/420) 29 and Yamamoto et al in Japanese patients (49.5%, 91/194) 30.
We identified eleven TP53 variants in our patients including ten Single Nucleotide Variants (SNV) and one Copy Number Variation (CNV). Among these variants, four have been newly identified in this study. They were not reported in any variant database (ClinVar, dbSNP, Varsome…). Effect prediction by SIFT and PolyPhen tools showed possible clinical significance of 2 missense variants on p53 function (c.62A>C (p.Asp21Ala); c.773A>T (p.Glu258Val)). Further studies are required to assess their functional significance.
The other seven variants identified in this study have been reported in Clinvar, dbSNP and gnomAD databases. Two of them are the most frequent (p.Pro72Arg, rs1042522) and the 16 bp insertion (rs59758982). They are both cancer related-variant and their functional impact in p53 have been reported 14. The first one (p.Pro72Arg, rs1042522) has been extensively studied as a potential risk factor for the development of malignancies and is the most common variant associated with predisposition to oral cancers. Allelic and genotypic distribution of rs1042522 are known depending on geographic latitude and ethnicity throughout the world 31, 32, 33, 34, 35. The variant has been associated in case/control studies with a higher risk of oral, nasopharyngeal, lung, thyroid, skin, cervical, prostate, bladder, gastric, colorectal, and hepatic cancers 20, 36, 37, 38, 39. Arg72 allele frequency was estimated at 31.26% in our patients, similar to frequencies observed in African and African American populations as reported in gnomAD database (31.4% and 32.1% respectively). In contrast, frequencies reported for Asian and Caucasian populations are higher (58.4% and 73.6% respectively) confirming the ethnic and geographic bias observed in the distribution of this variant throughout the world.
For the 16 bp insertion in intron 3 (rs59758982), its implication on carcinogenesis has also been highlighted in several cancers: colorectal, breast, cervical, gastric, head and neck, lung, bladder, esophagus, and in glaucoma 20, 40, 15. Allelic frequency of 12% has been reported in Caucasian diagnosed with colorectal cancer 40 whereas in our study, the frequency was 2 times higher (26.25%). In contrast this variant was not detected in 425 Japanese patients with primary open angle glaucoma 41.
Association study between these two cancer-related variants did not find any significant association in our population as we previously reported in a meta-analysis 19. However, LD test showed that they are strongly linked and could be transmitted together in a common haplotype predisposing towards oral cancers. It is therefore necessary to study the haplotype structure and investigate the role of neighboring variants spanning intron 3 to intron 4 region, especially intron 4 where an internal promoter encoding nine p53 isoforms has been located. Mutations in this promoter have been reported to affect the transcription and expression level of p53 isoforms 20, 42. Eiholzer and colleagues have recently demonstrated in 2020 that this genomic region harbor several haplotypes blocks that could increase the level of expression of the Δ133_TP53 transcript, which at certain levels of expression can have pro-tumor effects 43. Therefore it would be of interest to investigate by long read target sequencing and functional analysis this genomic region in patients diagnosed with cancer, and definitely address its implication on carcinogenesis.
Conclusion
Our study highlighted that despite the absence of association between the two most common cancer-related variants of TP53 observed in Senegalese patients diagnosed with oral cancer, their strong LD suggests that they are transmitted together in a common haplotype and may be implicated in oral carcinogenesis. Further functional and genomic studies are needed to explore the implication of the genomic region spanning intron 3 to intron 4 of TP53 gene where most cancer-related variants are located.
References
- 1.Bray F, Ferlay J, Soerjomataram I, R L Siegel, L A Torre.(2018)Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries.CA. , Cancer J 68(6), 394-424.
- 2.C P Freier, Stiasny A, Kuhn C, Mayr D, Alexiou C.. (2016)Immunohistochemical Evaluation of the Role of p53 Mutation in Cervical Cancer: Ser-20 p53-Mutant Correlates with Better Prognosis.Anticancer Res.36(6): 3131-7.
- 3.K R Patel, B N Vajaria, R D Singh, Begum R, P S. (2018) implications of p53 alterations in oral cancer progression: a review from India.Exp Oncol.40(1):. 10-18.
- 4.Stiasny A, C P Freier, Kuhn C, Schulze S, Mayr D.. (2017)The involvement of E6, p53, p16, MDM2 and Gal-3 in the clinical outcome of patients with cervical cancer.Oncol Lett.14(4): 4467-4476.
- 5.Choi S, J N Myers.(2008)Molecular pathogenesis of oral squamous cellcarcinoma:implications for therapy.J Dent Res.87(1):. 14-32.
- 6.Denaro N, Lo Nigro C, Natoli G, E G Russi, Adamo V.. (2011)TheRoleof p53 and MDM2 in Head and Neck Cancer.ISRN Otolaryngol.2011: 931813.
- 8.L G Entiauspe, F K Seixas, E M Nunes, F M Rodrigues, O A Dellagostin.. (2014)Uncommonnon-oncogenicHPVgenotypes, TP53 and MDM2genespolymorphismsin HIV-infectedwomeninSouthernBrazil.Braz J Infect Dis.18(6): 643-50.
- 9.Gupta S, Kumar P, B C Das.. (2018)HPV: Molecular pathways and targets.Curr Probl Cancer.42(2): 161-174.
- 10.Singh N, Sarkar J, K V Sashidhara, Ali S, Sinha S. (2014) activity of a novel coumarin-chalcone hybrid is mediated through intrinsic apoptotic pathway by inducing PUMA and alteringBax/Bcl-2 ratio.Apoptosis.19(6):. 1017-28.
- 11.Zushi Y, Narisawa-Saito M, Noguchi K, Yoshimatsu Y, Yugawa T.(2011)An in vitromultistepcarcinogenesismodel forbothHPV-positive and -negativehumanoralsquamouscellcarcinomas.Am. , J Cancer 1(7), 869-81.
- 12.Monti P, Menichini P, Speciale A, Cutrona G, Fais F.. (2020)Heterogeneityof TP53 Mutations and P53ProteinResidualFunctioninCancer:DoesItMatter?Front Oncol.10: 593383.
- 13.Alvarado-Ortiz E, G de la Cruz-Lopez K, Becerril-Rico J, M A Sarabia-Sanchez, Ortiz-Sanchez E.. (2020)Mutant p53 Gain-of-Function:Rolein CancerDevelopment, Progression, andTherapeuticApproaches.Front Cell Dev Biol.8: 607670.
- 14.Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G.. (2016)TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data.Hum Mutat.37(9): 865-76.
- 15.Gemignani F, Moreno V, Landi S, Moullan N, Chabrier A.(2004)A TP53polymorphismisassociatedwithincreasedriskof colorectal cancer andwithreducedlevelsof TP53mRNA.Oncogene.23(10):. 1954-6.
- 16.Barnoud T, Parris J L D, M E.(2019)Common genetic variants in the TP53 pathway and their impact on cancer.J Mol Cell Biol.11(7):. 578-585.
- 17.M E, Liu S, Yao S, Huo D, Liu Q.(2017)A functionally significant. SNP in TP53 and breast cancer risk in African-American women.NPJ Breast Cancer.3: 5.
- 18.Scheckenbach K, Lieven O, Gotte K, Bockmuhl U, Zotz R.(2004)p53 codon 72 polymorphic variants, loss of allele-specific transcription, and human papilloma virus 16 and/or 18 E6 messenger RNAexpression in squamous cell carcinomas of the head and neck.Cancer Epidemiol Biomarkers. Prev.13(11 Pt 1): 1805-9.
- 19.P D Sarr, S A Ba, Touré S, Diop J P D, DIA Y.. (2020)Association of the TP53 Arg72ProPolymorphismwithOralSquamousCellCarcinoma:A Meta-Analysis.J Mol Genet Med.14(3) .
- 20.Bellini I, Pitto L, M G, Porcu L, Moi P. (2010) expressionlevelsin relation to haplotypes of the TP53internalpromoterregion.Hum Mutat.31(4):. 456-65.
- 21.S J Diskin, Capasso M, Diamond M, D A Oldridge, Conkrite K.. (2014)Rare variants in TP53 and susceptibility to neuroblastoma.J Natl Cancer Inst.106(4): 047.
- 22.Ndiaye R, Dem A, P M Mbaye, P M Gueye, Diop G.(2014)[Study of codon 72 of p53 gene as a risk-factor in cervical cancer in Senegal].Bull Cancer.101(9):. 789-94.
- 23. (2021) Abbara A.Statistiques médicales et épidémiologiques : Outil de calcul médico-statistique permettant l'évaluation des indicateurs de risque et la liaison entre un facteur d'exposition et une maladie.2021. Available from:http://www.aly-abbara.com/utilitaires/statistiques/khi_carre_rr_odds_ratio_ic.html
- 24.Touré S, Sonko L, Diallo B-K, Diop R, Diop A.(2005)Profil épidémiologique des cancers de la cavité buccale au Sénégal.Rev Stomatol Chir Maxillofac.17.
- 25.F R Pires, A B Ramos, J B Oliveira, A S Tavares, P S Luz. (2013) casesfroma single oralpathologyserviceduringan 8-yearperiod.J Appl Oral Sci.21(5):. 460-7.
- 26.Rivera C, Venegas B. (2014) and molecular aspects of oral squamous cell carcinoma (Review).Oncol Lett.8(1):. 7-11.
- 27.Ara N, Atique M, Ahmed S, G Ali Bukhari S. (2014) of p53 gene mutation and protein expression in oral squamous cell carcinoma.J Coll Physicians Surg Pak.24(10):. 749-53.
- 28.E A Martinez, Jiménez-Gomez R, Medina C M A.(2013)Immunoexpressionof p53in Oral Squamous Cell Carcinoma and Oral Dysplastic Lesions in Patients with the Habit of Reverse Smoke.Int. , J 7(2), 7.
- 29.M L Poeta, Manola J, M A Goldwasser, Forastiere A, Benoit N.(2007)TP53 mutations and survival in squamous-cell carcinoma of the head and neck.N. , Engl J 357(25), 2552-61.
- 30.Yamamoto Y, Kanai M, Kou T, Sugiyama A, Nakamura E.(2020)Clinicalsignificanceof TP53 variants as possiblesecondaryfindingsintumor-onlynext-generationsequencing.J Hum Genet.65(2):. 125-132.
- 31.Beckman G, Birgander R, Sjalander A, Saha N, P A Holmberg. (1994) p53 polymorphism maintained by natural selection?Hum Hered.44(5):. 266-70.
- 32.Brant O, Hoffmann M, Kanappilly A, Gorogh T, Gottschlich S. (2007) codon 72polymorphismin squamous cell carcinoma of the head and neck region.Anticancer Res.27(5A):. 3301-5.
- 33.Fuentes M, Pulgar I, Gallo C, M C Bortolini, Canizales-Quinteros S.(2014)[Gene geography of Chile: regional distribution of American. European and African genetic contributions].Rev Med Chil.142(3): 281-9.
- 34.Kashima T, Makino K, Soemantri A, Ishida T.. (2007)TP53 codon 72polymorphismin 12 populations of insular Southeast Asia and Oceania.J Hum Genet.52(8): 694-7.
- 35.Sjalander A, Birgander R, Saha N, Beckman L, Beckman G. (1996) polymorphisms and haplotypes show distinct differences between major ethnic groups.Hum Hered.46(1):. 41-8.
- 36.Isakova J, Talaibekova E, Aldasheva N, Vinnikov D, Aldashev A.(2017)The association of polymorphic markers. Arg399Gln of XRCC1 gene, Arg72Pro of TP53 gene and T309G of MDM2 gene with breast cancer in Kyrgyz females.BMC Cancer.17(1): 758.
- 37.M S Khan, A, S R Masoodi, S H Khan, T A Rather.(2015)Significant association of TP53 Arg72Pro polymorphism in susceptibility to differentiated thyroid cancer.Cancer Biomark.15(4):. 459-65.
- 38.J Z Peng, Xue L, D G Liu, Y H Lin.. (2015)Association of the p53 Arg72Pro polymorphism withesophagealcancer in Chinesepopulations:a meta-analysis.Genet Mol Res.14(3): 9024-33.
- 39.Zhang Y, Zhang D, Zhao L, Sun L, Dong Q.. (2018)Associationbetweenp53 Arg72Propolymorphismand colorectal cancerriskin Asianpopulation:ameta-analysis.Curr Probl Cancer.42(6): 582-592.
- 40.Bottari F, Landi S, Gemignani F. (2006) tube genotyping of GSTM1. GSTT1 and TP53 polymorphisms by multiplex PCR.DNA Seq.17(5): 396-9.
- 41.Mabuchi F, Sakurada Y, Kashiwagi K, Yamagata Z, Iijima H.(2009)Lack of association between p53 gene polymorphisms and primary open angle glaucoma in the Japanese population.Mol Vis.15:. 1045-9.
Cited by (2)
This article has been cited by 2 scholarly works according to:
Citing Articles:
Cells (2023) OpenAlex
Cells (2023) Crossref
Dada Oluwaseyi Temilola, H. Adeola, Johan Grobbelaar, M. Chetty - Cells (2023) Semantic Scholar
